Molecular Models: Exploring the Structure of tRNA
- What is tRNA? - An introduction to the molecule
- Build a paper model of tRNA - Template and instructions for making the paper model
- Explore the atomic structure of tRNA - Interactive display of the atomic model of tRNA and details about its structure
1. What is tRNA?
|
During protein synthesis in cells, transfer RNA (tRNA) "translates" the genetic code in the messenger RNA (mRNA) into the language of proteins. Each tRNA molecule binds to a specific amino acid on the acceptor arm, recognizes its corresponding codon in the mRNA through the anticodon loop region, and delivers the amino acid to a growing peptide chain in the ribosome for protein synthesis. To learn more, see the Molecule of the Month features on tRNA and Ribosome. |

|
2. Build a Paper Model of tRNA
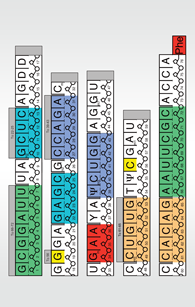
Use this PDF to build a paper model of the tRNA. As you build the model, you will see the primary structure, secondary structure and a simplified version of the tertiary structure of tRNA.
The tRNA is a polymer which has alternating sugar and phosphate units forming a backbone for the entire molecule. Each sugar in the backbone has a base attached to it which in turn interacts with other bases in the molecule.
The yeast phenylalanine tRNA has 76 bases which provide points of interaction between various parts of the molecule. In addition to standard bases such as adenine (A), cytosine (C), uracil (U) and guanine (G), tRNA has several modified bases such as pseudouridine (Ψ) and ribothymidine (T). These modifications are a hallmark of tRNA molecules and have been studied very carefully.
Although many of the details are still unknown, the modified bases are thought to tune the function of the tRNA, for instance, in optimizing the interaction of the anticodon with the mRNA codon.
Folding Instructions
Follow the instructions in the slide show to create your own model.
For additional help, a video demonstrating how to build this tRNA model is on YouTube.

-
Cut out each piece, leaving the grey tabs intact.

-
Following the nucleotide numbering, tape the pieces into a long strip. The blank grey tabs should be hidden.

-
Make slits in-between each double line (16 total). Be careful not to cut through the entire strip.

-
Fold back ("mountain fold") on the horizontal dashed lines.

-
The long strip shows the primary structure of tRNA.

-
Fold in ("valley fold") on the small vertical dotted lines.

-
Bring the blue (diagonally striped) bases 10-13 and 25-22 together and tape so the grey tab is hidden.

-
Bring the purple (vertically striped) bases 27-31 and 43-39 together and tape so the grey tab is hidden.

-
Bring the orange (triangle-patterned) bases 49-53 and 65-61 together and tape so the grey tab is hidden.

-
Bring the green (dotted) bases 1-7 and 72-66 together and tape.

-
At this point the model is a cloverleaf shape-the secondary structure of tRNA. Each colored region represents the double helical regions of the structure. Notice the almost-perfect base pairing (G:C and A:U bases).

-
Bring together bases G19 and C56 (colored yellow) to form a base pair - hide the grey tab by taping it under.

-
This forms the beginnings of the tRNA tertiary structure-the inverted L-shape. To further explore the tertiary structure of this tRNA, look at the Jmol model below.


Model limitations
In this paper model, base pairing between G19 and C56 is used to show the tertiary structure of tRNA. However, the actual molecule has many more interactions between these and other bases that stabilize the tertiary structure. These interactions can not be easily shown in the paper model. Explore the atomic model of tRNA to view in detail some of these interactions.3. Explore the Atomic Structure of tRNA
The atomic structure of tRNA can be visualized using coordinates from the Protein Data Bank. Here the structure from PDB ID 4tna is shown in a Jmol interactive view. Use the buttons to change the color and representation, and look closely at a few unusual representations between bases. For example bases C13, G22, m7G46 display a base triplet interaction (highlighted here as 13-22-46). Another unusual interaction occurs where base G57 stacks between the base pairs G18:(Ψ)55 and G19:C56 (highlighted here as 18-19 and 55-57).
Topics for further exploration
- Many different types of base pairs are formed when tRNA folds. These include typical A-U and C-G base pairs in the four stems, and some more unusual interactions when the whole thing folds into the final L shape. Use the model to find a non-standard base pair in one of the four stems, and use the Jmol to explore some unusual base interactions in the folded tRNA.
- What is the sequence of the anticodon in this tRNA? What codon would it read?