Staphylococcus aureus and Antibiotic Resistance
Staphylococcus aureus is a sphere-shaped bacterium that can cause infections in the skin, throat, and digestive tract.
Penicillin Binding Protein
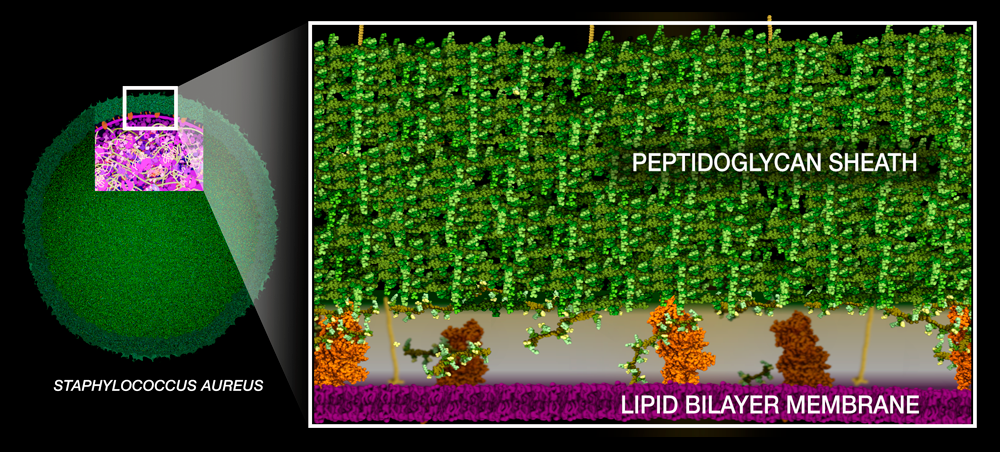
Like any gram-positive bacteria, Staph aureus builds a thick peptidoglycan sheath around its lipid bilayer membrane to protect itself from osmotic pressure.The building blocks of peptidoglycan consist of two sugars connected to a short chain of amino acids ending with two D-alanines with pentaglycine bridge extending to the side of the peptide. They are synthesized in the cytoplasm and then moved across the lipid bilayer membrane.
The building blocks are then assembled into chains via sugars, and the chains are crosslinked via the pentaglycine bridges. These processes are catalyzed by enzymes called penicillin-binding proteins (PBPs). The video below shows the crosslinking process catalyzed by the PBP (orange, PDB entry 5hl9).
Penicillin Action
The penicillin binding proteins, as the name suggests, have high affinity to bind penicillin and other beta-lactam antibiotics. The name beta-lactam antibiotics is derived from the core part of their chemical structure, the beta-lactam ring. Beta-lactam antibiotics bind covalently to the active serine residue in the PBPs, interrupting the crosslinking process and leaving the bacteria with an incomplete cell wall. As a result, bacteria become permeated with water and eventually explode.
Mechanisms of Resistance
In the face of overuse and misuse of antibiotics, new strains of Staphylococcus aureus have emerged that can proliferate even in the presence of the newest antibiotics. The resistance to beta-lactam antibiotics is possible due to 2 mechanisms:
1. Inactivation by a bacterial enzyme, beta-lactamase
Beta-lactamases (green, PDB entry 1ghp) are enzymes expressed by resistant bacteria that can cleave the beta-lactam ring in antibiotics, rendering them ineffective. In the presence of beta-lactamases, PBPs can continue their cross-linking activity and bacteria can thrive.
2. Alteration to the target site
The Methicillin-resistant Staphylococcus aureus (MRSA) is a strain of Staphylococcus aureus that developed mutations in the PBPs drug binding site (PBP2a, orange, PDB entry 1mwr) that are of no consequence to the enzyme's crosslinking activity, but render them immune to beta-lactam antibiotics.